An Introduction to Support Vector Machine (SVM) and the Simplified SMO Algorithm
3.00/5 (6 votes)
An introduction to the SVM and the simplified SMO algorithm
Introduction
In machine learning, support vector machines (SVMs) are supervised learning models with associated learning algorithms that analyze data used for classification and regression analysis (Wikipedia). This article is a summary of my learning and the main sources can be found in the References section.
Support Vector Machines and the Sequential Minimal Optimization (SMO) can be found in [1],[2] and [3]. Details about a simplified version of the SMO and its pseudo-code can be found in [4]. You can also find Python code of the SMO algorithms in [5] but it is hard to understand for beginners who have just started to learn Machine Learning. [6] is a special gift for beginners who want to learn about Support Vector Machine basically. In this article, I am going to introduce about SVM and a simplified version of the SMO by using Python code based on [4].
Background
In this article, we will consider a linear classifier for a binary classification problem with labels y (y ϵ [-1,1]) and features x. A SVM will compute a linear classifier (or a line) of the form:

With f(x), we can predict y = 1 if f(x) ≥ 0 and y = -1 if f(x) < 0. And by solving the dual problem (Equation 12, 13 in [1] at the References section), f(x) can be expressed:
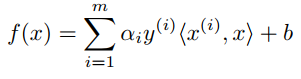
where αi (alpha i) is a Lagrange multiplier for solution and <x(i),x> called inner product of x(i) and x. A Python version of f(x) maybe look like this:
fXi = float(multiply(alphas,Y).T*(X*X[i,:].T)) + b
The Simplified SMO Algorithm
The simplified SMO algorithm takes two α parameters, αi and αj, and optimizes them. To do this, we iterate over all αi, i = 1, . . . m. If αi does not fulfill the Karush-Kuhn-Tucker conditions to within some numerical tolerance, we select αj at random from the remaining m − 1 α’s and optimize αi and αj. The following function is going to help us to select j randomly:
def selectJrandomly(i,m):
j=i
while (j==i):
j = int(random.uniform(0,m))
return j
If none of the α’s are changed after a few iterations over all the αi’s, then the algorithm terminates. We must also find bounds L and H:
- If y(i) != y(j), L = max(0, αj − αi), H = min(C, C + αj − αi)
- If y(i) = y(j), L = max(0, αi + αj − C), H = min(C, αi + αj)
Where C is a regularization parameter. Python code for the above is as follows:
if (Y[i] != Y[j]):
L = max(0, alphas[j] - alphas[i])
H = min(C, C + alphas[j] - alphas[i])
else:
L = max(0, alphas[j] + alphas[i] - C)
H = min(C, alphas[j] + alphas[i])
Now, we are going to find αj is given by:

Python code:
alphas[j] -= Y[j]*(Ei - Ej)/eta
Where:

Python code:
Ek = fXk - float(Y[k])
eta = 2.0 * X[i,:]*X[j,:].T - X[i,:]*X[i,:].T - X[j,:]*X[j,:].T
If this value ends up lying outside the bounds L and H, we must clip the value of αj to lie within this range:

The following function is going to help us to clip the value αj:
def clipAlphaJ(aj,H,L):
if aj > H:
aj = H
if L > aj:
aj = L
return aj
Finally, we can find the value for αi. This is given by:

where α(old)j is the value of αj before optimization. A version of Python code can look like this:
alphas[i] += Y[j]*Y[i]*(alphaJold - alphas[j])
After optimizing αi and αj, we select the threshold b:

where b1:

and b2:

Python code for b1 and b2:
b1 = b - Ei- Y[i]*(alphas[i]-alphaIold)*X[i,:]*X[i,:].T - Y[j]*(alphas[j]-alphaJold)*X[i,:]*X[j,:].T
b2 = b - Ej- Y[i]*(alphas[i]-alphaIold)*X[i,:]*X[j,:].T - Y[j]*(alphas[j]-alphaJold)*X[j,:]*X[j,:].T
Computing the W
After optimizing αi and αj, we can also compute w that is given:

The following function helps us to compute w from αi and αj:
def computeW(alphas, dataX, classY):
X = mat(dataX)
Y = mat(classY).T
m,n = shape(X)
w = zeros((n,1))
for i in range(m):
w += multiply(alphas[i]*Y[i],X[i,:].T)
return w
Predicted Class
We can predict which class a point belongs to from w and b:
def predictedClass(point, w, b):
p = mat(point)
f = p*w + b
if f > 0:
print(point," is belong to Class 1")
else:
print(point," is belong to Class -1")
The Python Function for the Simplified SMO Algorithm
And now, we can create a function (named simplifiedSMO) for the simplified SMO algorithm based on pseudo code in [4]:
Input
- C: regularization parameter
- tol: numerical tolerance
- max passes: max # of times to iterate over α’s without changing
- (x(1), y(1)), . . . , (x(m), y(m)): training data
Output
α : Lagrange multipliers for solution
b : threshold for solution
def simplifiedSMO(dataX, classY, C, tol, max_passes):
X = mat(dataX);
Y = mat(classY).T
m,n = shape(X)
# Initialize b: threshold for solution
b = 0;
# Initialize alphas: lagrange multipliers for solution
alphas = mat(zeros((m,1)))
passes = 0
while (passes < max_passes):
num_changed_alphas = 0
for i in range(m):
# Calculate Ei = f(xi) - yi
fXi = float(multiply(alphas,Y).T*(X*X[i,:].T)) + b
Ei = fXi - float(Y[i])
if ((Y[i]*Ei < -tol) and (alphas[i] < C)) or ((Y[i]*Ei > tol)
and (alphas[i] > 0)):
# select j # i randomly
j = selectJrandom(i,m)
# Calculate Ej = f(xj) - yj
fXj = float(multiply(alphas,Y).T*(X*X[j,:].T)) + b
Ej = fXj - float(Y[j])
# save old alphas's
alphaIold = alphas[i].copy();
alphaJold = alphas[j].copy();
# compute L and H
if (Y[i] != Y[j]):
L = max(0, alphas[j] - alphas[i])
H = min(C, C + alphas[j] - alphas[i])
else:
L = max(0, alphas[j] + alphas[i] - C)
H = min(C, alphas[j] + alphas[i])
# if L = H the continue to next i
if L==H:
continue
# compute eta
eta = 2.0 * X[i,:]*X[j,:].T - X[i,:]*X[i,:].T - X[j,:]*X[j,:].T
# if eta >= 0 then continue to next i
if eta >= 0:
continue
# compute new value for alphas j
alphas[j] -= Y[j]*(Ei - Ej)/eta
# clip new value for alphas j
alphas[j] = clipAlphasJ(alphas[j],H,L)
# if |alphasj - alphasold| < 0.00001 then continue to next i
if (abs(alphas[j] - alphaJold) < 0.00001):
continue
# determine value for alphas i
alphas[i] += Y[j]*Y[i]*(alphaJold - alphas[j])
# compute b1 and b2
b1 = b - Ei- Y[i]*(alphas[i]-alphaIold)*X[i,:]*X[i,:].T -
Y[j]*(alphas[j]-alphaJold)*X[i,:]*X[j,:].T
b2 = b - Ej- Y[i]*(alphas[i]-alphaIold)*X[i,:]*X[j,:].T
- Y[j]*(alphas[j]-alphaJold)*X[j,:]*X[j,:].T
# compute b
if (0 < alphas[i]) and (C > alphas[i]):
b = b1
elif (0 < alphas[j]) and (C > alphas[j]):
b = b2
else:
b = (b1 + b2)/2.0
num_changed_alphas += 1
if (num_changed_alphas == 0): passes += 1
else: passes = 0
return b,alphas
Plotting the Linear Classifier
After having alpha, w and b, we can also plot the linear classifier (or a line). The following function is going to help us to do this:
def plotLinearClassifier(point, w, alphas, b, dataX, labelY):
shape(alphas[alphas>0])
Y = np.array(labelY)
X = np.array(dataX)
svmMat = []
alphaMat = []
for i in range(10):
alphaMat.append(alphas[i])
if alphas[i]>0.0:
svmMat.append(X[i])
svmPoints = np.array(svmMat)
alphasArr = np.array(alphaMat)
numofSVMs = shape(svmPoints)[0]
print("Number of SVM points: %d" % numofSVMs)
xSVM = []; ySVM = []
for i in range(numofSVMs):
xSVM.append(svmPoints[i,0])
ySVM.append(svmPoints[i,1])
n = shape(X)[0]
xcord1 = []; ycord1 = []
xcord2 = []; ycord2 = []
for i in range(n):
if int(labelY[i])== 1:
xcord1.append(X[i,0])
ycord1.append(X[i,1])
else:
xcord2.append(X[i,0])
ycord2.append(X[i,1])
fig = plt.figure()
ax = fig.add_subplot(111)
ax.scatter(xcord1, ycord1, s=30, c='red', marker='s')
for j in range(0,len(xcord1)):
for l in range(numofSVMs):
if (xcord1[j]== xSVM[l]) and (ycord1[j]== ySVM[l]):
ax.annotate("SVM", (xcord1[j],ycord1[j]), (xcord1[j]+1,ycord1[j]+2),
arrowprops=dict(facecolor='black', shrink=0.005))
ax.scatter(xcord2, ycord2, s=30, c='green')
for k in range(0,len(xcord2)):
for l in range(numofSVMs):
if (xcord2[k]== xSVM[l]) and (ycord2[k]== ySVM[l]):
ax.annotate("SVM", (xcord2[k],ycord2[k]),(xcord2[k]-1,ycord2[k]+1),
arrowprops=dict(facecolor='black', shrink=0.005))
red_patch = mpatches.Patch(color='red', label='Class 1')
green_patch = mpatches.Patch(color='green', label='Class -1')
plt.legend(handles=[red_patch,green_patch])
x = []
y = []
for xfit in np.linspace(-3.0, 3.0):
x.append(xfit)
y.append(float((-w[0]/w[1])*xfit - b[0,0])/w[1])
ax.plot(x,y)
predictedClass(point,w,b)
p = mat(point)
ax.scatter(p[0,0], p[0,1], s=30, c='black', marker='s')
circle1=plt.Circle((p[0,0],p[0,1]),0.6, color='b', fill=False)
plt.gcf().gca().add_artist(circle1)
plt.show()
Using the Code
To run all of Python code above, we need to create three files:
- The myData.txt file contains training data:
-3 -2 0 -2 3 0 -1 -4 0 2 3 0 3 4 0 -1 9 1 2 14 1 1 17 1 3 12 1 0 8 1
In each row, the two first values are features and the third value is a label.
- The SimSMO.py file contains functions:
def loadDataSet(fileName): dataX = [] labelY = [] fr = open(fileName) for r in fr.readlines(): record = r.strip().split() dataX.append([float(record[0]), float(record[1])]) labelY.append(float(record[2])) return dataX, labelY # select j # i randomly def selectJrandomly(i,m): ... # clip new value for alphas j def clipAlphaJ(alphasj,H,L): ... def simplifiedSMO(dataX, classY, C, tol, max_passes): ... def computeW(alphas,dataX,classY): ... def plotLinearClassifier(point, w, alphas, b, dataX, labelY): ... def predictedClass(point, w, b): ... - Finally, we need to create the testSVM.py file to test functions:
import SimSMO X,Y = SimSMO.loadDataSet('myData.txt') b,alphas = SimSMO.simplifiedSMO(X, Y, 0.6, 0.001, 40) w = SimSMO.computeW(alphas,X,Y) # test with the data point (3, 4) SimSMO.plotLinearClassifier([3,4], w, alphas, b, X, Y)
The result can look like this:
Number of SVM points: 3
[3, 4] is belong to Class -1
And:

Points of Interest
In this article, I only introduced the SVM basically and a simplified version of the SMO algorithm. If you want to use SVMs and the SMO in a real world application, you can discover more about them in documents below (or maybe more).
References
- [1] CS229 Lecture notes, Andrew Ng, Support Vector Machines
- [2] Bingyu Wang, Virgil Pavlu, Support Vector Machines
- [3] John C. Platt, Fast Training of Support Vector Machines using Sequential Minimal Optimization
- [4] CS 229, Autumn 2009, The simplified SMO Algorithm
- [5] Peter Harrington, Machine learning in Action
- [6] Alexandre Kowalczyk, Support Vector Machines Succinctly
History
- 18th November, 2018: Initial version
