In the Finite Element Analysis, sparse matrices have a particular form - the diagonals are non-zero, and each row has a predictable upper bound on non-zero entries. Research codes often don't care about efficiency, but for large geometries the performance impact is measurable.
Introduction
The Finite Element Method (FEM) was conceived as an engineering tool for calculating forces in trusses.
It allowed engineers to "see" forces that are invisible to the naked eye.
It was quickly realized that aside from analyzing static force distributions,
FEM can be applied in various areas of physics - Fluid Mechanics, Heat Transfer, Electromagnetism.
In general, FEM is a way of "solving" differential equations by converting them to something
that a computer "understands" - floating-point arithmetic.
While there are various types of FEM, many of them come down to crunching a linear system
of equations in the form
$\begin{aligned} \mathbf{Ax}=\mathbf{b}, \end{aligned}$
where \(\mathbf{A}\) is a sparse matrix.
Libraries that solve such equations already exist
- MKL, LAPACK, cuSPARSE - to name a few. FEM codes prepare the matrix \(\mathbf{A}\) and the
right-hand side \(\mathbf{b}\) and let a solver do its magic.
But how exactly is the sparse matrix prepared? Is there a way to optimize this process?
During my several years in academia, I wrote an algorithm for the assembly of sparse matrices.
This data structure is unlike any other. It is a non-standard mapping with specific properties. Most attempts
to go with the "standard" hash sets and red-black trees go terribly wrong.
This article will discuss the possible pitfalls with the assembly of sparse linear systems.
It may be helpful to researchers who develop custom FEM implementations.
A Simple Example
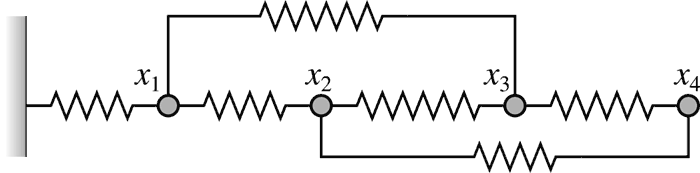
Consider a mass-spring system, where each spring has its unique stiffness constant
\(k_n\) and a rest length \(d_n\).
A spring \(n\) that connects nodes \(i\) and \(j\)
exerts the following force onto node \(i\).
$\begin{aligned} F_{n,i} = k_n(x_j-x_i-d_n).\end{aligned}$
For every force acting on node \(i\),
there is a force with an opposite sign acting on the incident node \(j\):
$\begin{aligned} F_{n,j}= -F_{n,i}.\end{aligned}$
We are looking for the nodal displacements at equilibrium - when all forces from the springs
add up to zero:
$\begin{aligned} \sum_{n} {F_{n,k}} = 0.\end{aligned}$
A linear system of equations arises:
$\begin{aligned} \mathbf{Ax}=\mathbf{b}. \end{aligned}$
where \(\mathbf{x}=[x_1,x_2,...,x_N]^T\) is the nodal positions vector,
\(\mathbf{A}\) is a matrix of nodal interactions, and
\(\mathbf{b}\) is the right-hand side of the linear system.
The latter is non-zero because
the initial spring lengths \(d_n\) contribute constant terms.
The length of \(\mathbf{x}\) will be denoted as N,
and the size of matrix \(\mathbf{A}\) is NxN.
For each spring that connects the nodes \(i\) and
\(j\), there are non-zero entries
\(e_{ii},e_{ij},e_{ji},e_{jj}\) in matrix
\(\mathbf{A}\). Aside from these elements, the
matrix is mostly filled with zeros - it is sparse.
This article will discuss
the ways of preparing and storing sparse matrices for FEM applications.
COO List - Super Easy

Most solver libraries accept sparse matrices in the so-called Coordinate List
(COO)
format, which represents the non-zero entries as
(row, column, value) tuples. Row and column
are encoded as integers, whereas value entries have the floating-point type.
To create such a list of tuples, one would iterate over all springs and append the
corresponding entries to the list.
std::vector<std::tuple<int,int,double>> COO_list;
for(const auto &spring : springs)
{
COO_list.insert(A.end(),spring.Entries_A());
}
This can be accomplished in parallel with OpenMP and TBB containers:
tbb::concurrent_vector<std::tuple<int,int,double>> COO_list;
#pragma omp parallel for
for(unsigned i=0;i<springs.size();i++)
{
const Spring &spring = springs[i];
A.push_back(spring.Aii());
A.push_back(spring.Aij());
A.push_back(spring.Aji());
A.push_back(spring.Ajj());
}
There are a few problems with this approach.
- Since all threads append the entries simultaneously
to the same container, there are frequent race conditions.
Concurrent containers work best when the race conditions are infrequent.
- Some
(row,column) pairs may appear
more than once. The solver will subsequently add them together, but they take more memory than needed. - For solvers, COO is not a convenient format,
so they will spend time converting it to a more suitable representation.
Despite the obvious limitations, this assembly method is probably the best
tradeoff between code complexity
and performance. It is definitely a good option, especially for research codes
that do not push the RAM/CPU limits.
Flawed Options that are Worse than COO
In systems with more than a million variables, the assembly step may take a significant amount of time. Unlike
the one-dimensional spring model, which creates four entries per spring, some FEM elements may create much larger entries.
For example, a cohesive zone element may connect two quads, i.e., affect eight nodes. Additionally, each node may have three
degrees of freedom (xyz), making each sub-entry of size 24x24, giving a total of 576 double-precision values.
Storing all entries in a list may affect performance and consume more memory than needed.
Since each row of the matrix contains at least one non-zero entry, the matrix is dense
in its rows. One may try to store it as a vector of vectors of pairs:
std::vector<std::vector<std::pair<int,double>>> crazy_vector;
Some sparse matrix libraries use this approach because it allows the insertion
of new non-zero entries by dynamically resizing the arrays. This storage format
also facilitates iterating over row entries, which simplifies matrix multiplication algorithms.
The problem is that none of these benefits speed up the assembly process. Re-allocation
of the vectors leads to memory fragmentation and overall performance loss,
while insertion operations on vectors are super slow.
Addressing the Problem
Sparse matrix libraries provide on-the-fly resizing, but for most
FEM applications, this is not needed. Since we know which elements connect to which nodes, we can infer
the locations of non-zero entries upfront before their values are computed.
This brings us to the
Compressed Sparse Row (CSR) format, which
is a native internal representation in many solvers. Submitting this format to a solver will not
trigger a conversion step. Unlike other representations, it is not suitable for dynamic resizing,
but it is efficient for storage and access.

In CSR, non-zero entries are stored sequentially in a single allocated block.
Before that block is allocated, the
"sparsity structure" is created, i.e., the list of non-zero entries should be known.
For the mass-spring example, the code would look something like this:
std::vector<std::pair<int,int>> s;
for(const auto &spring : springs)
{
s.emplace_back(spring.i, spring.j);
s.emplace_back(spring.j, spring.i);
}
for(int i=0;i<N;i++) structure.emplace_back(i,i);
std::sort(s.begin(),s.end());
s.resize(std::distance(s.begin(),std::unique(s.begin(), s.end())));
unsigned NNZ = s.size();
std::vector<double> values;
values.resize(NNZ);
The resulting structure vector s contains one (i,j)-entry
for each non-zero element of the sparse matrix. The size of s is
the number of non-zero elements, denoted as NNZ.
This approach looks very similar to the first attempt with the COO list.
We are simply creating the
COO list without the actual values.
The algorithm marks which (i,j) cells
are in use, removes the duplicates and prepares a single memory block
for the values.
But now the values are in a separate list!
Why are two lists better than one? Well, the assembly stage can now be truly parallel.
Race conditions will occur when two springs write into the same (i,j)-cell.
Such a situation, while not impossible, is very unlikely.
So the assembly stage has improved. A typical implementation would look like this:
std::fill(values.begin(), values.end(), 0);
#pragma omp parallel for
for(unsigned i=0;i<springs.size();i++)
{
const Spring &spring = springs[i];
#pragma omp atomic
values[get_offset(spring.i, spring.i)] += spring.Aii();
#pragma omp atomic
values[get_offset(spring.i, spring.j)] += spring.Aij();
#pragma omp atomic
values[get_offset(spring.j, spring.i)] += spring.Aji();
#pragma omp atomic
values[get_offset(spring.j, spring.j)] += spring.Ajj();
}
Mappings, Mappings...
We now need a mapping function get_offset(int i, int j)
that points to where each (i,j)-entry is stored. A naive solution would be
to use std::map or std::unordered_map,
but let's consider the downsides.
- These data structures allocate memory dynamically every time they are prepared.
- The insertion complexity is O(log(NNZ)), so for several million
NNZs
there will be a perceivable performance hit. - Sorting (or maintaining the sorted order) of
the
(i,j)-list is computationally expensive.
Final Solution
Let's use the fact that our sparse matrix
is dense in terms of rows. We know that each row contains at least one
non-zero entry, and the number of rows N is known upfront. Thus,
(i,j)-pairs can be represented as a vector of vectors (again).
constexpr unsigned nCon = 8;
std::vector<std::unique_ptr<tbb::concurrent_vector<unsigned>>> rows;
if(rows.size()<N)
{
rows.reserve(N);
while(rows.size()<N)
rows.push_back(std::make_unique<tbb::concurrent_vector<unsigned>>(nCon*2));
}
We are back to where we started, but not quite. This time the situation is different.
We are not dynamically allocating the whole matrix but only the structure arrays.
In fact, we avoid unnecessary reallocations altogether.
The maximal number of entries per row is more or less known upfront -
it depends on a particular FEM algorithm and the mesh type. Still, it is usually constant
within a given application. For example, in 2D meshes
with triangular elements, each node is typically surrounded by 6-7 triangles.
Storing this upper bound in nCon constant ensures that the row vectors
will not exceed their initial capacity. Additionally, this whole memory structure can be reused
on subsequent computation steps, and allocation only happens once.
Finally, since the TBB concurrent vectors are used, the structure can be populated in parallel.
I have not compared the sequential and parallel algorithms, but the
parallel is probably faster for extra-large meshes, while
the sequential is optimal for smaller geometries.
For the sequential algorithm, one could use Boost's small vector
boost::container::small_vector<unsigned,nCon*2>.
Let's see how the non-zero elements are marked.
#pragma omp parallel for
for(unsigned k=0;k<springs.size();k++)
{
const Spring &spring = springs[k];
rows[spring.i]->push_back(j);
rows[spring.j]->push_back(i);
}
Instead of sorting one ginormous array of (i,j)-indices,
we now sort many
small arrays in parallel! The duplicate
(i,j)-entries are removed as before.
Finally, we count the total number of
nonzero elements NNZ,
which will be needed in the next step.
unsigned NNZ = 0;
#pragma omp parallel for reduction(+:NNZ)
for(unsigned i=0;i<N;i++)
{
tbb::concurrent_vector<unsigned> &rn = *rows[i];
rn.push_back(i);
std::sort(rn.begin(),rn.end());
rn.resize(std::distance(rn.begin(),std::unique(rn.begin(),rn.end())));
NNZ += rn.size();
}
We now proceed with creating the structure arrays
COL_INDEX and ROW_INDEX, which
are a part of the CSR representation and passed to the solver.
They also allow an O(1) lookup in the (i,j)->(offset) mapping.
The size of COL_INDEX is NNZ, and the size of ROW_INDEX is
N+1. We store both arrays as std::vector<int> and resize as needed. To populate them,
we iterate over the sorted (i,j)-pairs and enumerate the entries sequentially.
std::vector<int> COL_INDEX, ROW_INDEX;
COL_INDEX.resize(NNZ);
ROW_INDEX.resize(N+1);
ROW_INDEX[N] = NNZ;
unsigned count = 0;
for(unsigned row=0;row<N;row++)
{
ROW_INDEX[row] = count;
tbb::concurrent_vector<unsigned> &rn = *rows[row];
for(unsigned const &local_column : rn)
{
COL_INDEX[count] = local_column;
count++;
}
}
Last Piece of the Puzzle
At the assembly stage, the matrix entries are additively written into the values
array. For each (i,j)-entry, there is an offset to a memory location.
It is important to infer these offsets efficiently because this operation repeats
numerous times. In the earlier versions, I tried various approaches, starting from a straightforward
std::map<std::pair<int,int>,int>, then evolving into per-row maps, and even
a separate vector of vectors. But is there a better way?
There is an elegant way to use COL_INDEX and ROW_INDEX.
The latter already stores the
offsets to the first non-zero element in each row, ROW_INDEX[i].
We just need to find the column-index that matches j.
We iterate through
a portion of COL_INDEX between &COL_INDEX[ROW_INDEX[i]] and
&COL_INDEX[ROW_INDEX[i+1]], and find the value that matches j.
unsigned get_offset(int row, int column)
{
int col_offset_begin = ROW_INDEX[row];
int col_offset_end = ROW_INDEX[row+1];
int *start_pt = &COL_INDEX[col_offset_begin];
int *end_pt = &COL_INDEX[col_offset_end];
auto it = std::lower_bound(start_pt, end_pt, column);
return std::distance(start_pt,it)+col_offset_begin;
}
I am not exactly sure whether the binary search std::lower_bound
or a regular search std::find is faster in this case.
Depending on a particular FEM setup, rows may have very few entries,
so a direct linear search may perform better. Either way, this
is an elegant solution with constant time lookup.
Conclusion
There are numerous varieties of Finite Element Methods. Students and researchers
are constantly inventing new modifications. Not all FEM codes use sparse matrices -
some can do without them. This article does not claim to suggest the
most optimal algorithm - it's and algorithm that works, and there is room for optimizations.
In my Ph.D. and Postdoc research, I wrote various FEM codes -
materials with cohesive zones, shell and plate elements, 2D Neo-Hookean materials, among other things.
Some used linear solvers, others used quadratic optimizers.
My focus was more on FEM formulations rather than the implementation of the matrix math.
Indeed, some competitive programmers would quickly come up with an optimal assembly procedure,
but for me, this code was slowly evolving into its current shape over many years.
A working project the uses this algorithm can be found
here. Please
do not hesitate to leave a comment, especially to suggest improvements. This algorithm
may have practical value for other FEM researchers, and that's the motivation for
this article!
I am a researcher working on numerical models for deformation, crushing and flow of ice. The models are based on continuum mechanics, where numerical approaches include particle-based methods and finite elements.
 General
General  News
News  Suggestion
Suggestion  Question
Question  Bug
Bug  Answer
Answer  Joke
Joke  Praise
Praise  Rant
Rant  Admin
Admin